Commercial vs. Academic Software
/0 Comments/in DBS/by andreashornRecently, I had the pleasure to meet with an R&D representative of a major DBS electrode manufacturer and we talked about software built to model deep brain stimulation electrodes in computer simulations, such as Lead-DBS.
This meeting – and the development of Lead-DBS since 2012 – made me think about pros and cons of academic vs. commercial software with similar goals. A few aspects were new to me and could be of interest, so I decided to write them up.
Since a few years, software that aims at modeling DBS electrodes in computer simulations has been emerging across the globe, both authored by research institutions and industry. On the academic side, Cicerone DBS (1) was a pioneering platform developed by the McIntyre lab later bought by Boston Scientific and turned into the commercial application GUIDE. Around the same time, the Yelnik/Bardinet group in Paris developed the deformable YeB atlas that went into the commercial Optivize (2) software by Medtronic and is now used in it’s successor, SureTune 3. The company Brainlab also develops similar software with some more focus on the pre-surgical / planning part. CranialCloud, developed by the spinoff Neurotargeting LLC out of Vanderbilt University also develops a product aimed at modeling DBS electrodes.
We started development of Lead-DBS in 2012 to fill a clinical need at our center, since at the time, no commercial or academic software was available for download purchase. Later on, the first similar software that was made publicly available was the AFNI based DBSproc. There are multiple other academic projects in development, such as PyDBS in Rennes, PaCER in Luxembourg, “VirtualPatient” at MGH in Boston, DBSmapping in Porto, StimVision at Case Western – this list may not be exhaustive. At the point of writing, the only two academic toolboxes that are openly available for download are DBSproc and Lead-DBS (3).
The obvious
A few obvious pros and cons for commercial vs. academic software exist: Of course, academic software is most often free and open source. It may be customized, you are free to plug in your own research scripts and modify everything tailor-cut to your needs. For some researchers, this is a massive advantage. I would even argue that some research projects may just not be feasible with commercial software since it’s often hard to automate, batch-process or interface with these tools.
On the other hand, commercial software is shiny, easy to use, comparably bug free and if need be, you can often call a hotline or even a sales-rep for in-house support. These advantages can be massive, as well. Hassle-free work with a product in a pipeline that just needs to function is often not easily imaginable with academic software. In a purely clinical setting, where not-so tech-savvy residents need to quickly localize electrodes in the brains of their patients can easily become a nightmare with academic and their home-brew nature.
Keeping pace with research
A slightly less obvious advantage of research software is that it is much more flexible. Commercial applications undergo tedious certification processes to achieve CE-marks or FDA-approvals and may safely be applied in clinical context. This is a great advantage for clinicians but drastically slows down development of software. New research developments may not be easily integrated into commercial tools since they need to be certified, too. On the wild-west side of academic software, however, new tools can be integrated from idea & concept to end-user deployment within days. For instance, in 2009, the global fibertracking approach (‘Gibbstracking, Reisert 2009’) won the Neurospin fibercup and was thus evaluated as the best fibertracking software compared to 9 competitors. We integrated this software into Lead-DBS when we started it. Recently, a new comparative study found the generalized Q-sampling algorithm implemented in DSI studio most promising. With some help from Fang-Cheng Yeh, the author of DSI studio, we were able to integrate this approach into Lead-DBS within a few weeks. Fibertracking is an extremely good example to highlight differences between academic and commercial pipelines – the latter often still apply a combination of tensor-based DTI & Mori Streamline tracking.
Another good example is a brainshift correction feature in Lead-DBS that developed from concept to published code on a 3-day hackathon at MIT. Such a pace is not imaginable for commercial applications – but naturally, it may also bear some risks.
Reproducibility and open science
Clinicians may not care too much about this and likely trade open source for a CE-mark / FDA approval in a snap. Still, using a commercial software without published methods does not fulfill criteria of reproducible science and is a no-go in the eyes of most researchers. Moreover, in the short history of DBS imaging suites, already three of them were announced to reach market shortly but soon after withdrawn, their development discontinued (4).
In the short period between announcement and withdrawal, some scientific colleagues had enthusiastically started research projects that would make use of the tools because they had been given exclusive pre-market access to the software. After withdrawal of the tools, reproducibility of their scientific findings was not guaranteed anymore.
Moreover, sales representatives of two major electrode manufacturers told me that their companies did not have any interest in publishing methods of the software packages (such as details about nonlinear warping techniques applied or the estimation of activation volumes around DBS electrodes).
Self-limitation
One reason I write this post is to highlight a thought that had not been obvious to me for some time. Namely, algorithms applied in commercial applications are often limited by strict time constrains. For instance, there are various methods to perform nonlinear multispectral co-registration between patient and template anatomy implemented in Lead-DBS. If one chooses the BSplineSyn approach included in Advanced Normalization Tools and selects the “fine” preset with additional subcortical refinement, a registration will use up ~50GB of RAM and it is really helpful to run it on a compute cluster. Lead-DBS proposes of an internal job submitting system that works on usual high performance clusters with some minor modifications. [Needless to say, Lead-DBS can easily be used on a standard laptop and most users do so – but then choose less costly deformation strategies]. In academic environments, processing time is of secondary importance (e.g. less important than accuracy of results). Scientists are used to long job walltimes and for most, it is not a dealbreaker if a job takes a few hours or a night. In contrast, this doesn’t fit into a clinical workflow where results are needed fast. An R&D representative of a company told me their product was limited to processing times below an hour, a few minutes were optimal. This is understandable given the target user group of their products – but it deliberately hinders their algorithms from realizing their full potential.
Conclusion
I hope this post could elaborate some pros and cons about commercial and academic software in the field of DBS imaging. It is not meant to discretize either/or – on the contrary, being a clinician and researcher myself, I am absolutely convinced that there is a strong need for both types of software. Still, I hope I could convince you about some issues in using either type of software for the wrong task. Needless to say, I’d love to hear your feedback or thoughts.
Footnotes
- to the best of our knowledge development discontinued but still available for macaques
- to the best of our knowledge development discontinued
- to the best of our knowledge
- referring to Boston Scientific GUIDE system, Medtronic OptiVize and the initial version of Medtronic SureTune
DBS targets in MNI space
/0 Comments/in DBS/by andreashornThis blog post summarizes the work in

The aim of this study was to transform surgical literature targets – that are conventionally reported relative to the anterior and posterior commissures of the brain – within MNI space. We will first quickly revise the definition of these two “standard stereotactic” spaces and then summarize the methodology of the study.
What are AC/PC coordinates?
Surgical coordinates defined relative to the anterior (AC), posterior commissure (PC) or their midpoint, the midcommissural point (MCP) are also called “functional coordinates” since they are used in functional neurosurgery (the clinical domain largely dominated by deep brain stimulation surgery).
This coordinate system was popularized by two seminal surgical atlases, the Schaltenbrandt-Wahren (1977) and Talairach-Tournoux (1988) atlases [as a side note, the idea of stereotactic coordinates first appeared as early as 1906 by Clarke & Horsley]. As pointed out by Brett and colleagues (2002), at their time, these atlases introduced three innovations: a coordinate system that helped to identify brain locations relative to anatomical landmarks, an atlas describing a standard brain, with anatomical labels and cytoarchitectonic details contributed by the work of Korbinian Brodmann (1925) and a 9-parameter transformation (adjusting for position, orientation and size) to match one brain to another. In general, the AC and PC are not the worst reference points given their relatively small anatomical variance across subjects. However, the two atlases were both defined based on single brains that were smaller than average. Moreover, as pointed out by Weiss and colleagues (2002), the reference points are not always the same – sometimes the center of the AC and PC are used, sometimes their anterior/posterior borders, sometimes a mixture of the two.
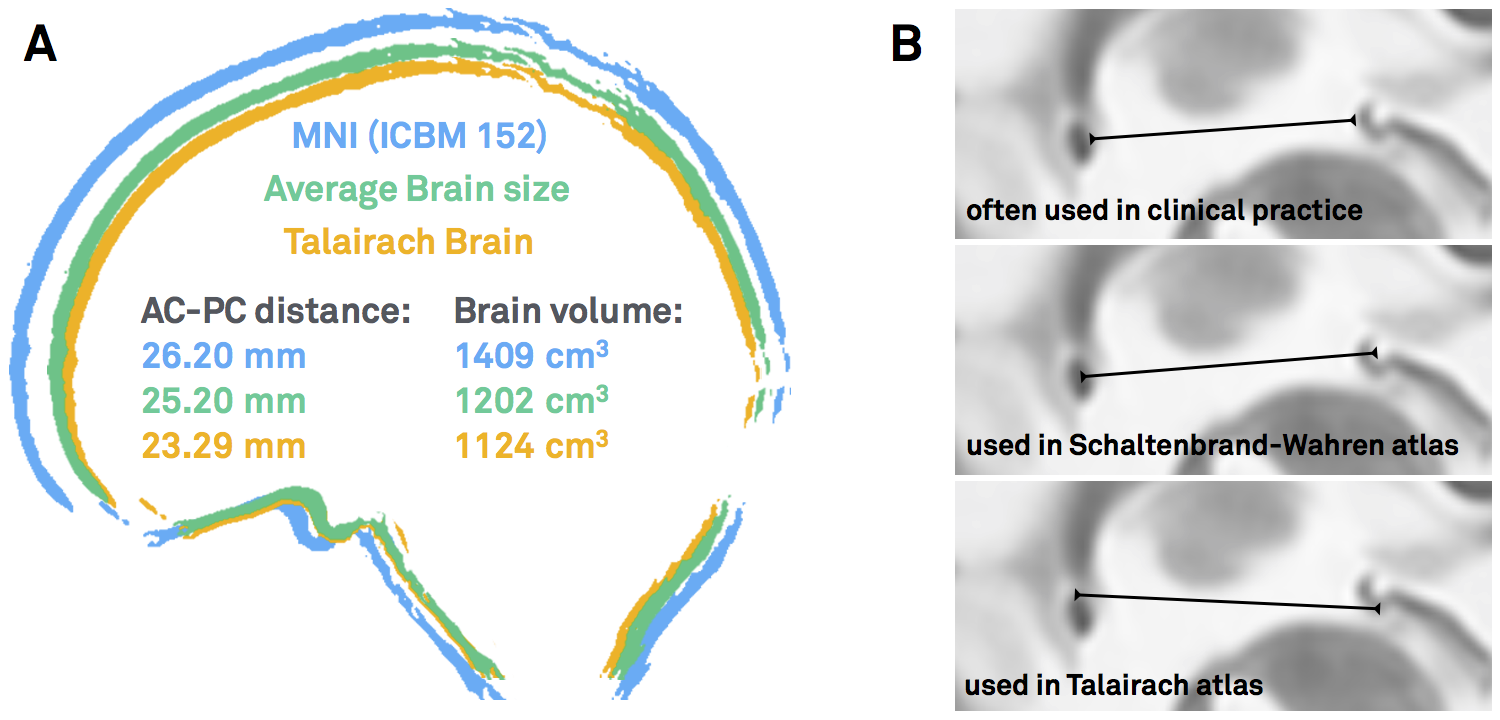
Figure 1: AC/PC-coordinates are ill-defined. A) Both the Schaltenbrandt-Wahren and Talairach-Tournoux atlases are based on single brain that are smaller than average. B) The reference points of the anterior and posterior commissures are not always defined exactly the same – depending on the atlas used and the surgeon’s preference.
What are MNI coordinates?
To cite Matthew Brett again, “the MNI wanted to define a brain that is more representative of the population. They created a new template that was approximately matched to the Talairach brain in a two-stage procedure. First, they took 241 normal MRI scans, and manually defined various landmarks, in order to identify a line very similar to the AC-PC line, and the edges of the brain. Each brain was scaled to match the landmarks to equivalent positions on the Talairach atlas. They then took 305 normal MRI scans (all right handed, 239 M, 66 F, age 23.4 +/- 4.1), and used an automated 9 parameter linear algorithm to match the brains to the average of the 241 brains that had been matched to the Talairach atlas. From this they generated an average of 305 brain scans thus transformed – the MNI305”
“The problem introduced by the MNI standard brains is that the MNI linear transform has not matched the brains completely to the Talairach brain. As a result the MNI brains are slightly larger (in particular higher, deeper and longer) than the Talairach brain.”
This quote highlights the problem of conversions between Talairach and MNI space which has been extensively studied in the imaging literature before (e.g. see pioneering work by Lancaster & colleagues).
However, surgical coordinates are not even in Talairach space.
…at least not always. As mentioned above, depending on the surgical target and their personal preference, surgeons choose to report their findings relative to the AC, the PC or the MCP. To add another source of confusion, coordinates may be reported in millimeters lateral, anterior and below the MCP. This seems to be the most frequent case, but does not always apply (coordinates could also be reported as lateral, posterior and above the AC). In contrast, Talairach coordinates are standardized to be always relative to the AC and the signs of each coordinate always make clear where one is in the brain (e.g. a negative x-coordinate means left hemisphere, a positive z-coordinate dorsal to the AC).
Thus, existing Talairach to MNI conversion tools only partially apply to the heterogeneous definition of coordinates expressed in the neurosurgical literature.
Why bother with MNI coordinates, anyways?
One could argue that the AC/PC system is established and there is no need to convert coordinates into MNI space altogether. However, the following reasons motivated us to still aim at creating a conversion tool that would to exactly that:
- As mentioned, traditional atlases still used in neurosurgical literature (and available “in AC/PC space”) are based on single brains and not representative of the average population
- Then, the number of these atlases is very limited. In contrast, a multitude of atlases have been defined in MNI space – and stem from various types of data such as histology, fMRI, diffusion MRI or meta analyses
- Beyond atlases, other resources – such as normative connectomes or whole-brain histology datasets are available in MNI space
- If a conversion tool existed, findings from neurosurgical and functional imaging literature could be combined
- Finally, the MNI space(s) are defined by detailed three-dimensional templates and nonlinear deformation algorithms have been trained across ~15 years of fMRI literature to warp single brains into their space and back. This means that once something is defined in MNI space, it may be transformed into the space of any other space, more or less accurately
[As a side note, the term “MNI space” itself is pretty ill-defined. For an explanation why, please read this blog post].
So how to do it?
I hope I could convince you that it may be a good idea to build a tool that may transform from AC/PC- to MNI cooordinates (and back). One problematic fact is that AC/PC coordinates are expressed relative to the AC/PC of a specific (clinical) population – and the specific anatomy of this population is often unknown.
Clearly, the optimal way would be to obtain MRIs from the specific population, mark their AC and PC fiducials, mark the specific coordinate of interest and nonlinearly warp this coordinate into MNI space. However, obtaining MRIs from large clinical cohorts may render difficult if not impossible.
Instead, we used surrogate populations to do the same thing. Two DBS cohorts with different surgical targets, from which all coordinates were known, served as a gold standard. Then, we defined their AC/PC coordinates in healthy young, age-matched, disease-matched and disease-severity matched surrogate cohorts to warp their AC/PC coordinates into MNI space.
Notably, this was done in a probabilistic fashion: A single point (relative to AC/PC) could end up in different places if defined in different brains. Defining this point in various brains and warping each of them to MNI space results in a probabilistic point cloud.
Our main finding was that an age-matched surrogate cohorts yielded similar results compared to the actual cohort. Only using a Young/Healthy cohort would render inferior results; a disease- or even disease-severity-matched cohort was not needed.
The conversion tool applied
Once we showed that it was feasible to convert AC/PC coordinates into MNI space, we performed a literature review of effective stereotactical diseases in MNI space. For each study that reported an effective DBS target, we gathered an age-matched cohort from the IXI dataset. We then used this surrogate-cohort to probabilistically warp the AC/PC coordinate into MNI space.
The result was an atlas of DBS targets for various diseases defined in MNI space
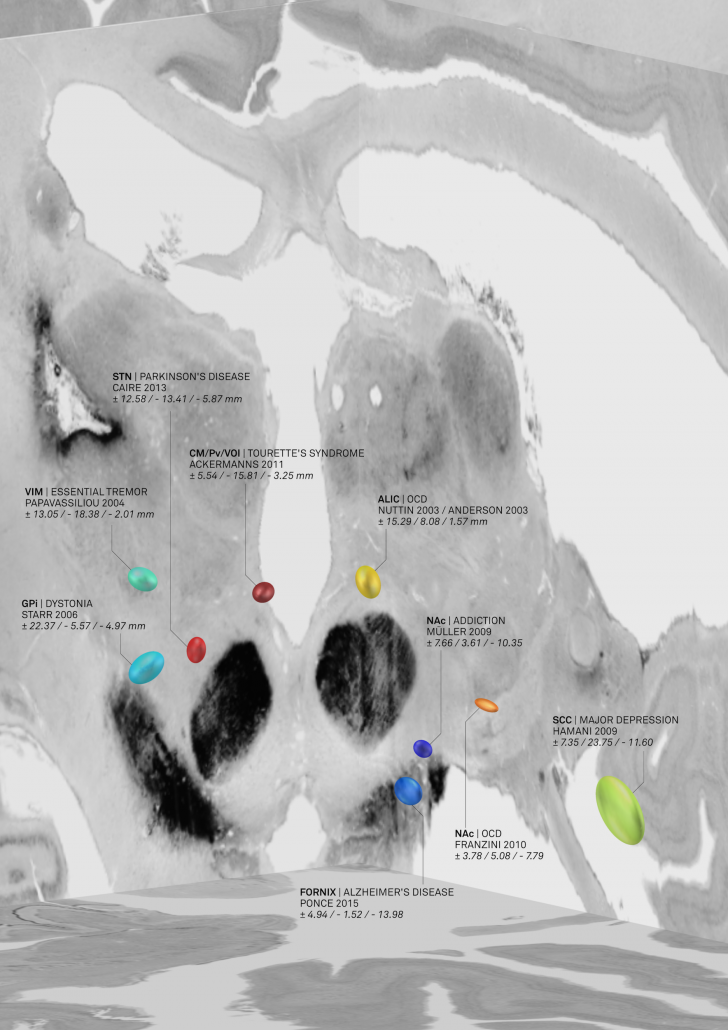
Figure 2: AC/PC coordinates transformed to MNI space based on age-matched surrogate cohorts. This picture was elected for coverart in NeuroImage Volume 150/June 2017 and directly shows the combination of findings with «MNI resources». Namely, the backdrop of the image is defined by the BigBrain dataset, a whole-brain histology stack that has been warped into MNI.
The nice thing about these results is that we could now compare the DBS target sites with other subcortical atlases – i.e. not only with the Schaltenbrandt/Wahren atlas commonly used in the neurosurgical literature. For instance, the Morel or Chakravarty atlases are based on histology and available in MNI space:
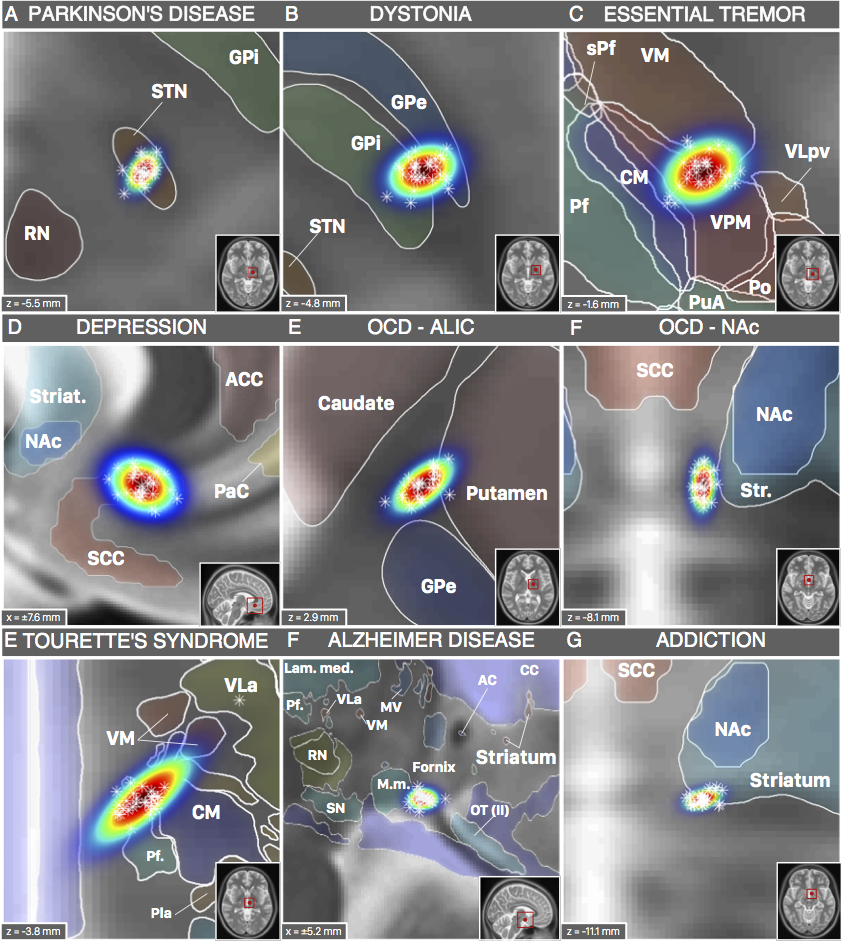
Figure 3: DBS targets defined in MNI space. Each target was mapped to MNI space using a healthy cohort that was age-matched to the cohort in which the AC/PC coordinate had been defined. Various atlases available in MNI space can help to interpret anatomical context.
However, not only structural atlases are available as «ressources in MNI space». We aimed at making this point by characterizing the spatial position of the DBS target for Essential Tremor. This target is commonly assumed to reside within the ventrointermediate (VIM) nucleus of the thalamus. However, this assumption relies on early studies dating back to the pre-MRI era (e.g. by work of Hassler or Jones). Moreover, the nucleus is defined as VIM/Vim in the Walker/Ohye notations, respecitvely. It is called Vimi in the thalamic nomenclatures by Hassler and Van Buren & Borke and as vim in the Dewulf notation. In the popular Jones / Hirai & Jones notations, the VLpv or VPM best corresponds to the nucleus and in the nomenclatures by Ilinski/Kultas-Ilinsky and Percheron, the VL and LI/LIL nuclei best represent the Walker VIM.
Thus, depending on the atlas, the same nucleus will be called differently and not exhibit exactly the same borders. Moreover, the DBS target used to treat Essential Tremor has been reported to reside below the thalamus, in the subthalamic area/dorsal zona incerta. Furthermore, the dentatorubrothalamic tract seems to play a crucial role mediating therapeutic effect.
Finally, thalamic atlases exist that are not based on histology but on functional zones in the thalamus defined by functional or diffusion-weighted imaging. For instance, the seminal work by Behrens and colleagues (2003) defined seven functional zones of the thalamus based on their structural connectivity. This was reported as a group result in MNI space and could thus now be combined with findings from the neurosurgical literature:
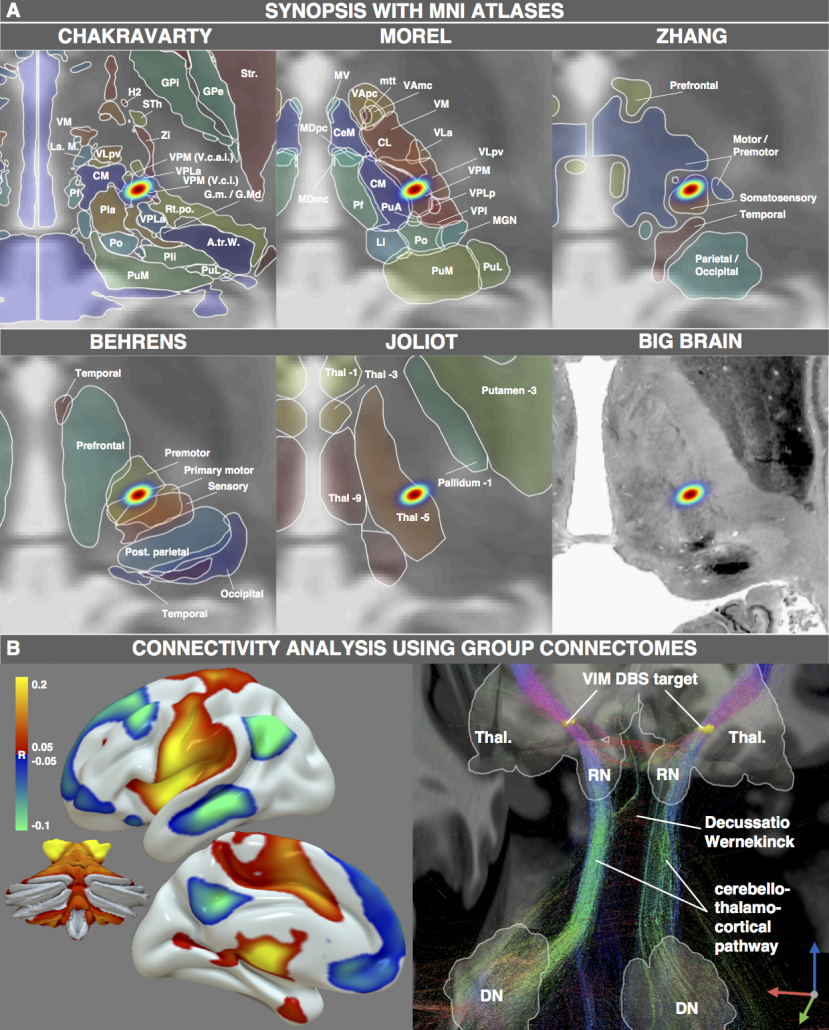
Figure 4: One exemplary DBS target (to treat Essential Tremor) extensively characterized by various MNI resources. Atlases based on histology, functional or diffusion-weighted MRI may be used to characterize the spatial location of the target. Moreover, normative functional and structural connectome datasets may be used to characterize the connectivity profile of the effective target site.
Figure 4 summarizes the potential and the gain in knowledge entailed with warping neurosurgical targets into MNI space. The methodology described in this article is fully implemented within Lead-DBS and can be applied to any AC/PC target based on a surrogate cohort of choice using the AC/PC <=> MNI conversion tool:
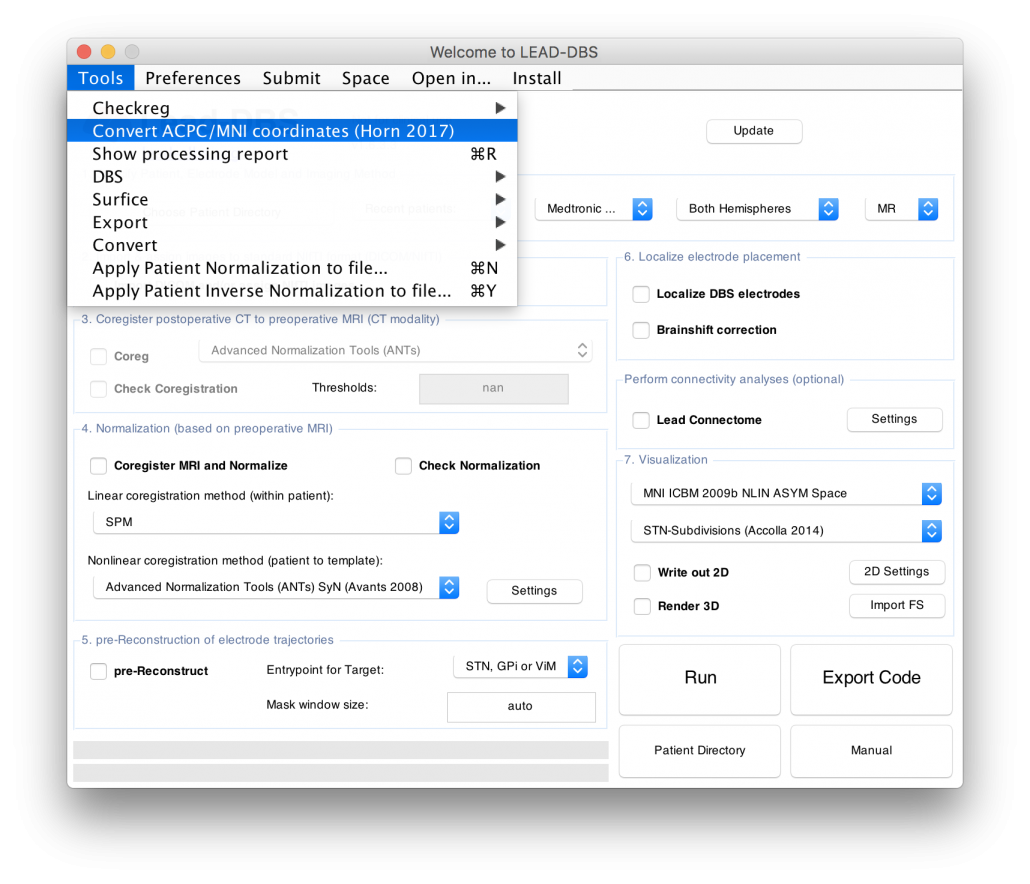
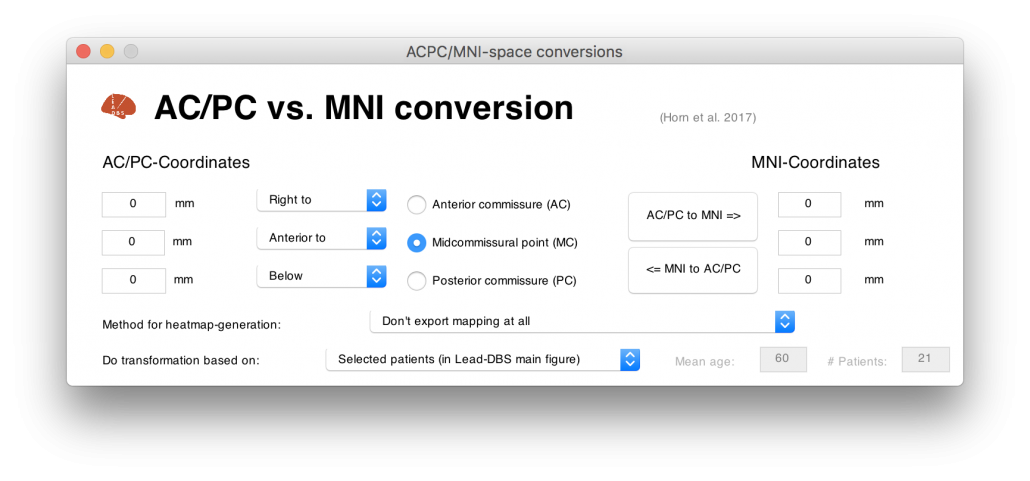
Read the full article here: Horn, A., Kühn, A. A., Merkl, A., Shih, L., Alterman, R., & Fox, M. (2017). Probabilistic conversion of neurosurgical DBS electrode coordinates into MNI space. NeuroImage, 150, 395–404.

