A while ago, Matthew Brett wrote a review paper about the “MNI space”, its historical development and origins which largely focused on the differences between Talairach and MNI space.
Since first publication of this post, a better resource about different coordinate systems has been established by the BIDS team.
Nowadays, the Talairach space doesn’t play a big role in modern neuroscience anymore, however, the MNI space of course continues to do. This post is motivated to shed some light onto the different “MNI spaces” available today, their differences and commonalities. It is known to some but should be known to more people in the field that there is no such thing as a single “MNI space”. This fact motivates this post.
Within the field of classical neuroimaging (fMRI, dMRI), the slight differences between various versions of the MNI space may not matter. However, in disciplines like deep brain stimulation imaging – where millimeters do matter – the subtle changes between spaces may definitely impact results.
A brief overview about the historical development (largely informed by the Brett post and paper)
- MNI305: 305 normal MRI brains were linearly coregistered (9-param) to 241 brains that had been coregistered (roughly) to the Talairach atlas. This was the first MNI template. It is T1 only.
- Colin27 original: Colin Holmes, a lab member of the MNI, was scanned 27 times. This was used as the standard template in SPM96. This template was matched to the MNI305. In the original release, only a T1 template was available.
- MNI152 linear: 152 normal images were linearly coregistered (9-param) to the 305 space. This was adopted to define standard anatomy by the International Consortium of Brain Mapping (ICBM) and is used by SPM from version 99 on. Only a T1 template is available.
- SPM: Note that in later iterations, SPM defined its own “space” based on scans acquired within the IXI dataset, leveraging on the concept of segmentation-based normalizations (as applied in the Unified Segmentation, DARTEL and SHOOT approaches). This space, referred to as the IXI549Space is largely similar to MNI152 and of similar resolution (where it’s likely safe to say that differences do not matter as much for most applications). Along the same lines, both spaces are unsuitable for use in DBS imaging, in our opinion. More info here.
- MNI152 NLIN 6th generation: Here, the 152 brains were nonlinearly registered into 305 space. Again, only a T1 template is available.
- FSL: The FSLR is an adapted version of this space but in its asymmetric flavor (MNI152NLIN6Asym, hence different to the official one provided here). The template also provides surface coordinates references. More info here.
- Colin27 hires T1/T2 version (2008): The MNI also released a high-resolution version of the Colin27 space.
- MNI152 NLIN 2009: Finally, in 2009, the MNI released an updated version after again nonlinearly co-registering the 152 acquisitions. This template exhibits the best resolution and detail to date and is available in T1, T2, PD and T2-relaxometry versions. It has not (yet) been adopted by SPM or FSL though and it’s anatomy is slightly different in direct comparison to the 6th generation templates. Moreover, it comes in three versions (two of which are explicitly mentioned here) that each have a symmetric and asymmetric subversion:
- MNI152 NLIN 2009a: Probably the official new template in 1 × 1 × 1 mm resolution. It comes in T1, T2, PD and T2-relaxometry versions. There is a symmetric and an asymmetric version (latter not featureing T2-relaxometry).
- MNI152 NLIN 2009b: The only of the novel templates available in 0.5 × 0.5 × 0.5 mm resolution. It is available in a T1, T2 and PD version. Due to the high resolution of this template, it has been adopted by Lead-DBS as the primary anatomy template. Lead-DBS uses the asymmetric version of this template.
- There are more MNI templates for different species and different cohorts.
- Within the world of Freesurfer, the fsaverage space has been roughly / approximately coregistered to MNI space (for more details see here, thanks to Denise Ruprai) but is based on different subjects (as far as I know based on the Buckner40 dataset which has become part of the genomics superstruct repository).
- Thomas Yeo’s lab has published a paper about accurate conversions between MNI and freesurfer space, code to do this is openly available here.
- Finally, the Minn/Wash-U Human Connectome Project nonlinearly registered 900 of their subjects into MNI 152 6th-generation space, i.e. into the space used by SPM/FSL. Their templates are high in detail and especially subcortically may exceed the signal to noise contrast of the MNI 2009 versions. They are – however – in a slightly different space as the 2009 versions (namely in 6th-gen space).
You can download all official MNI templates here. The HCP templates can be downloaded here.
Also see this page for further info on different spaces with additional information on which spaces SPM and FSL use.
Space or template – what’s the difference?
So what is “the MNI space” now, really? Generally speaking, the MNI “space” merely defines the boundaries around the brain, expressed in millimeters, from a set origin. Depending on which template you use, a certain coordinate may end up in a different anatomical structure. In many applications (e.g. fMRI on an average 3T scanner), this doesn’t matter too much. Standard research pipelines use smoothing (sometimes with FWHM kernels of 8 × 8 × 8 mm (!)) and fMRI resolution sometimes is as poor as 3 × 3 × 3 mm. These subtle differences between templates may not be a big problem. However, in DBS imaging, millimeters do matter and Lead-DBS has been optimized to account for that as good as possible. This is the reason why output folders of datasets processed with Lead-DBS are sometimes quite huge in size.
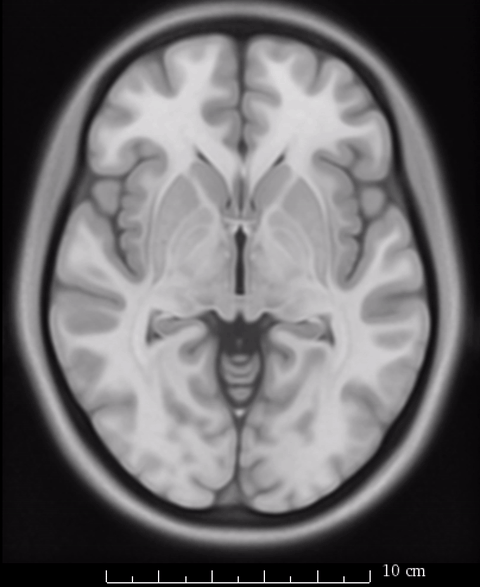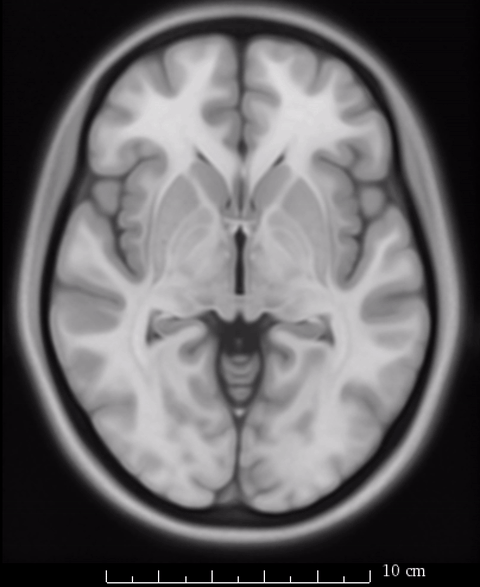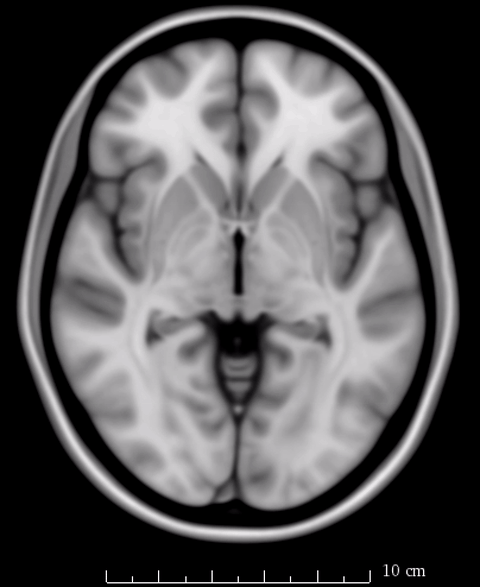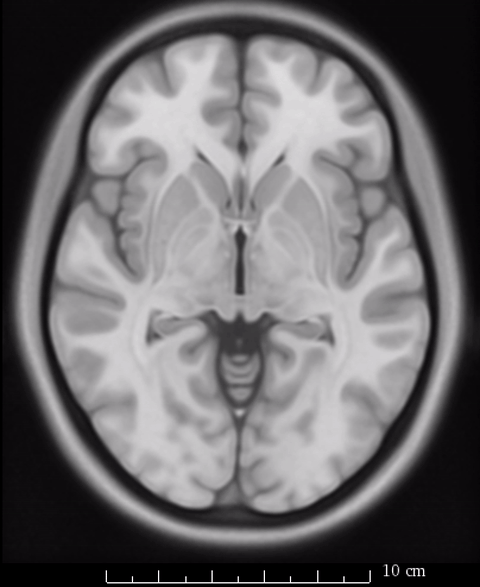 So rather of speaking about which “space” we report results in, we should refer to the specific template on which we report our results. As mentioned above, Lead-DBS generally uses the MNI 2009b NLIN asymmetric T2 template as it’s primary definition of default anatomy. However, not all resources and routines within the software have been or can be adapted to that exact space.
So rather of speaking about which “space” we report results in, we should refer to the specific template on which we report our results. As mentioned above, Lead-DBS generally uses the MNI 2009b NLIN asymmetric T2 template as it’s primary definition of default anatomy. However, not all resources and routines within the software have been or can be adapted to that exact space.
In contrast, within the literature, many papers refer to “standard space” or “MNI space” only. Depending on the context, based on this information, one may assume that they used either the linear or nonlinear 6th-gen 152 space T1 templates (since this is the one supplied with SPM or FSL). However, even if results are reported within “MNI 152 space”, one wouldn’t know which of the three 152 spaces is meant and if the 2009 series is used, which of the 6 template series (2009a, 2009b or 2009c? symmetric or asymmetric?) was used.
The image above fades between 6th gen (the slighly fuzzier image, used by SPM/FSL) and 2009b T1 (more precise image, used by Lead-DBS). On a whole-brain basis, this difference may seem minimal, but small subcortical structures such as the GPi and STN may move significantly depending on which template is being used.
The special case of Lead-DBS – what’s in 2009b NLIN, and what is not?
Unfortunately, not all resources are available within 2009b NLIN space. On the contrary, due to the fact that FSL and SPM still use the 6th-gen space templates, most MNI resources are indeed available in this space only. It was never an option to use the 6th-gen templates within Lead-DBS, however, since no T2-version of it exists, the most common DBS target is the STN and thus most research centers acquire T2-weighted acquisitions pre- and postoperatively.
Recently, with the help of Vladimir Fonov from the MNI, we estimated a nonlinear warp between 6th generation MNI T1 and 2009b NLIN space that is publicly available here (2). Even though this transform is based on T1 images only (again, no T2-version of the 6th gen templates exist), it yields accurate results.
From Lead-DBS v.1.4.9.4 on, we applied this transform to the following resources:
- Subcortical atlases (found under /lead_dbs/atlases/)
- ATAG_Linear (Keuken 2014)
- ATAG_Nonlinear (Keuken 2014)
- ATAG_STN (Forstmann 2012 & Keuken 2013)
- Oxford Thalamic Connectivity Atlas (Behrens 2003)
- BGHAT (Prodoehl 2008)
- CFA Subcortical Shape Atlas (Qiu 2010)
- Harvard AAN atlas (Edlow 2012)
- Templates (found under /lead_dbs/templates/)
- mni_hires_fa.nii (which is the FMRIB FA58 template)
What is perfectly “2009b safe” within Lead-DBS?
- In our view, the transformed atlases mentioned above are in good agreement with the 2009b template
- The DISTAL and Human Motor Thalamus Atlases were created for Lead-DBS and are inside the correct space.
- The Accolla STN subdivision atlas has been defined on the 2009 MNI space
- Nonlinear deformations using Advanced Normalization Tools (ANTs) within Lead-DBS are 100% 2009b conform (they use the template itself)
- SPM New Segment uses a segmentation of the ICBM2009b template performed based on the Lorio 2016 (3) enhanced TPM maps that have been graciously provided for use within Lead-DBS by Bogdan Draganski. Thus, this normalization routine perfectly conforms to the 2009b template.
- Similar to the above, DARTEL & SHOOT implementations within Lead-DBS use this novel TPM automatically built by Lead-DBS.
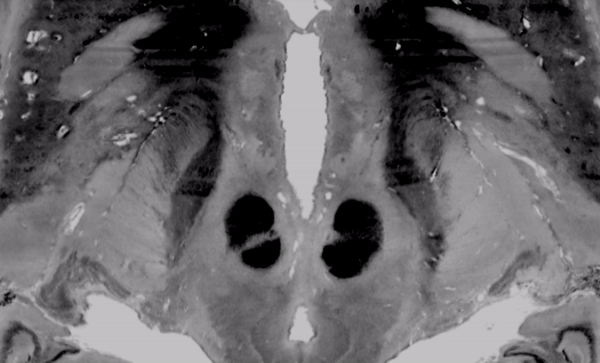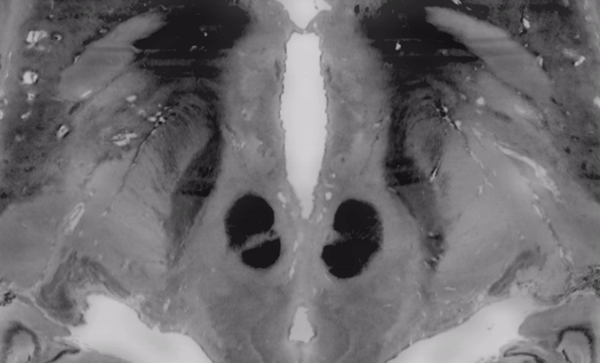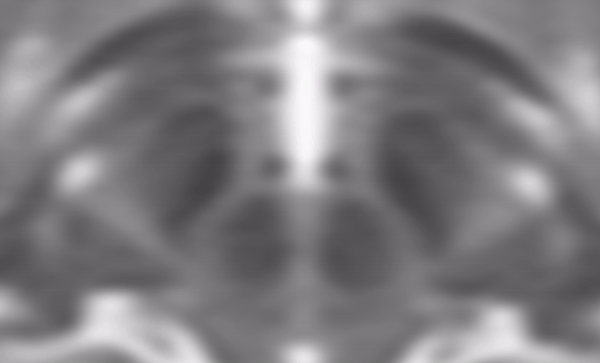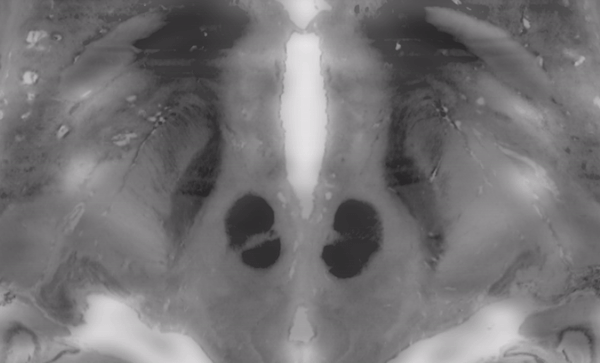
The above image faded between subcortical structures of the BigBrain 2015 release in 100um resolution and the MNI/ICBM 153 2009b NLIN asymetric template.
What is not “2009b safe” within Lead-DBS?
The following resources may not be exactly inside 2009b space:
- Atlases not mentioned in the above lists or atlases you added yourself to Lead-DBS. In case they had been originally co-registered to 6th generation templates, you may consider applying our transform (2).
- Whole-brain parcellation maps used for connectomic imaging (in lead_dbs/templates/labeling) have not been modified. We argue that the slight differences on cortical structures between 6th gen MNI space and 2009 MNI space will not yield significantly different results in whole-brain connectometric analyses and did not apply any transform to these files. In our view, they are still good to use.
We dearly hope that this article was useful and did not lead to more confusion. In general, differences between various MNI templates are subtle. Still, they can be meaningful depending on the research question so we think that a higher awareness of different templates within the research community is a good thing.
References
(1) Brett, M., Johnsrude, I. S., & Owen, A. M. (2002). The problem of functional localization in the human brain. Nature Reviews Neuroscience, 3(3), 243–249. http://doi.org/10.1038/nrn756
(2) Horn, Andreas (2016): MNI T1 6thGen NLIN to MNI 2009b NLIN ANTs transform. figshare. https://dx.doi.org/10.6084/m9.figshare.3502238.v1
(3) Lorio, S., Fresard, S., Adaszewski, S., Kherif, F., Chowdhury, R., Frackowiak, R. S., et al. (2016). New tissue priors for improved automated classification of subcortical brain structures on MRI. NeuroImage, 130, 157–166. http://doi.org/10.1016/j.neuroimage.2016.01.062

