Crucial methodological updates for Lead-DBS
We are thrilled that the Lead-DBS v2 methodological paper is out in NeuroImage.
[An unpaywalled preprint can be found here: https://www.biorxiv.org/content/early/2018/08/28/322008].
Lead-DBS was developed since 2012 and the first methods paper published in 2014. Since then, nearly everything has changed – and this led us to writing up the aforementioned updated methods paper. In the manuscript, we describe tools for multispectral nonlinear registration, structural/functional connectivity analyses, brain shift correction, reconstruction of microelectrode recordings and orientation detection of segmented DBS leads. The long author list reflects how many people have contributed to Lead-DBS development since then.
One particularly important processing step is nonlinear registration (patient brains to an atlas or the other way around). Currently, ten approaches to do so are readily built into Lead-DBS and each comes with adjustable settings. Thus, it became very important to investigate and quantify results that may originate from those different approaches. In a marathon effort spearheaded by Siobhan Ewert and led by Todd M Herrington that involved >11,000 nonlinear warps, the two most promising normalization approaches were identified – ANTs based multispectral SyN registration as well as multispectram SPM-based segmentation – which are now the default choices in Lead-DBS. In fact, automatic segmentations of the STN/GPi performed using those approaches were marginally if not at all significantly worse than manual expert segmentations.
Their article can be found here:
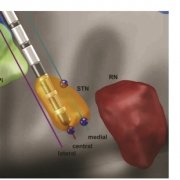
Finally, a new study out of Bern/Oxford validated the default STN atlas defined in Lead-DBS (Siobhan Ewert’s DISTAL atlas, Toward defining deep brain stimulation targets in MNI space:…) using microelectrode recordings. This important work showed that anatomical and electrophysiological accuracy was highest for the DISTAL atlas when comparing it to two different atlases of the STN. The article can be found here: