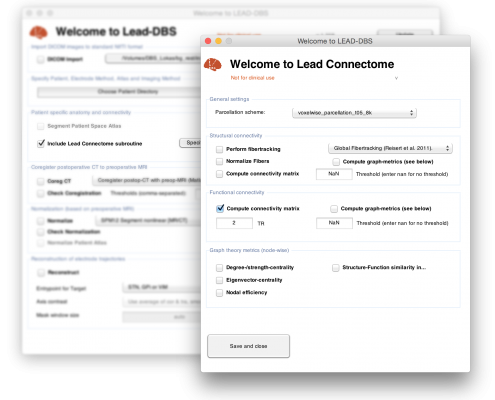
LEAD Connectome – as part of LEAD DBS – is a complete structural-functional connectomic analysis pipeline in MATLAB
You can use LEAD Connectome within but also completely outside the context of deep brain stimulation. Thus, LEAD Connectome can be used for connectomic analyses of
- healthy subjects
- patients
- patients that undergo DBS surgery.
The foundation of the code is based on the work published here:
The software can be used to perform structural and functional connectomic analyses which are based on
- dMRI (diffusion-weighted imaging based fiber tractography – often simply referred to as DTI) and
- rs-fMRI (resting-state functional magnetic resonance imaging) data respectively
Structural connectivity may be analyzed using the GQI method implemented in DSI Studio, the global “Gibbs’ tracking” method implemented in the DTI & Fibertools for SPM software package, which must be installed and on your Matlab path. Other tracking algorithms can be used, of course, too. As a simple example, a global tracking algorithm made by Dirk-Jan Kroon is pre-installed. The toolbox is easy to extend for implementation of your own fibertracking algorithm, if you prefer so.
Functional connectivity can be analyzed right out of the box (only SPM needs to be installed and on your search path).
As always, please use the forum for questions!